Algorithm Software Homer
Homer Software and Data Download HOMER Software for motif discovery and next-gen sequencing analysis HOMER Motif Analysis HOMER contains a novel motif discovery algorithm that was designed for regulatory element analysis in genomics applications (DNA only, no protein). It is a differential motif discovery algorithm, which means that it takes two sets of sequences and tries to identify the regulatory elements that are specifically enriched in on set relative to the other. Erfurt Luger Serial Numbers there. It uses ZOOPS scoring (zero or one occurrence per sequence) coupled with the hypergeometric enrichment calculations (or binomial) to determine motif enrichment. HOMER also tries its best to account for sequenced bias in the dataset.
It was designed with ChIP-Seq and promoter analysis in mind, but can be applied to pretty much any nucleic acids motif finding problem. There are several ways to perform motif analysis with HOMER.
The links below introduce the various workflows for running motif analysis. In a nutshell, HOMER contains two tools, findMotifs.pl and findMotifsGenom Email Tarantula Keygenguru. e.pl, that manage all the steps for discovering motifs in promoter and genomic regions, respectively. These scripts attempt to make it easy for the user to analyze a list of genes or genomic positions for enriched motifs. However, if you already have the sequence files that you want to analyze (i.e.
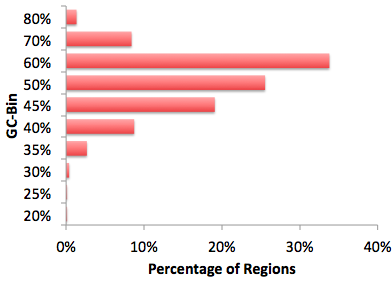
Free Criminal Lawyers In Sacramento there. FASTA files), findMotifs.pl (and homer2) can process these directly. Often the target sequences have an imbalance in the sequence content other than GC%. This can be caused by biological phenomenon, such as codon-bias in exons, or experimental bias caused by preferrential sequencing of A-rich stretches etc.